Pandas load data
Pandas is a a great package for data analysis. It is required by packages such as: xarray, seaborn, pymc3, plotnine, magenta, google-colab, featuretools, fastai, and others. In here we discuss how to load data into pandas dataframe.
Table of contents:
- Creating a dataframe
- Read textual dataframe
- When the load is slow
- Parse as a date type
- Read dataframe from the CSV file
- Read dataframe from remote csv file
- Read dataframe from HTML page
- Fixing missing values
- Convert string to categories
- What machine learning algorithms need?
- Fast save and load DataFrame in pandas
Creating a dataframe
Let’s create a dataframe in pandas. Simplified dataframe is a table. To create a table we just need the table size.
Example:
df = pd.DataFrame(index=range(14),columns=range(7))
print(df)
Output:
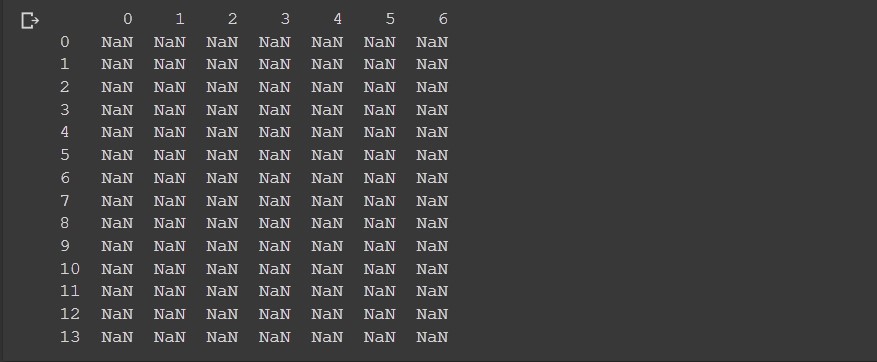
Here we set the 7 columns and 14 rows. Note how in python pandas the start index is always 0.
To create a dataframe you may also provide the column names and row index names.
Example:
import pandas as pd
df = pd.DataFrame(index=['a', 'b', 'c'], columns=['time', 'date', 'name'])
print(df)
print(df.values)
Output:
time date name
a NaN NaN NaN
b NaN NaN NaN
c NaN NaN NaN
[[nan nan nan]
[nan nan nan]
[nan nan nan]]
From the output when we printed print(df.values) we got something that looks like a list. What is this?
Actually if we check the type of type(df.values) object we will find this is numpy.ndarray.
Pandas requires numpy, python-dateutil and pytz packages. Confirm that with
pip show pandas.
Does this just mean we need numpy array to provide data to pandas dataframe?
No, because pandas is smart. Consider this example:
Example:
data = pd.DataFrame([[10, 11, 12, 13, 14],
[22, 23, 24, 25, 26],
[31, 32, 33, 34, 35]])
print(data)
Output:
0 1 2 3 4
0 10 11 12 13 14
1 22 23 24 25 26
2 31 32 33 34 35
In here we haven’t provided numpy arrays, instead the input data for the DataFrame is list of lists.
What will happen internally? Internally, pandas will convert the list of lists to numpy array.
Example:
import numpy as np
ll = [[10, 11, 12, 12, 14],
[22, 23, 21, 22, 23],
[31, 35, 32, 34, 34]]
npa = np.asarray(ll)
data = pd.DataFrame(npa)
print(data)
Another example:
data = pd.DataFrame({'Col1': [10, 11, 12, 13, 14],
'Col2': [22, 23, 24, 25, 26],
'Col3': [31, 32, 33, 34, 35]})
print(data)
Output:
Col1 Col2 Col3
0 10 22 31
1 11 23 32
2 12 24 33
3 13 25 34
4 14 26 35
If we plan to set the row names, this would be also possible:
Example:
data = pd.DataFrame({'Col1': [10, 11, 12, 13, 14],
'Col2': [22, 23, 24, 25, 26],
'Col3': [31, 32, 33, 34, 35]},
index=['row1', 'row2', 'row3', 'row4', 'row5'])
print(data)
Output:
Col1 Col2 Col3
row1 10 22 31
row2 11 23 32
row3 12 24 33
row4 13 25 34
row5 14 26 35
Read textual dataframe
Comma separator
First example will use the read_csv() function to read a multiline text. CSV means Comma Separated Values. Using csv format is very frequent in pandas.
Example:
import pandas as pd
import io
text=u"""asdf,333
asdf,444
asdf,555"""
df=pd.read_csv(io.StringIO(text),
sep=r',',
header=None,
engine='python',
encoding = "iso-8859-1")
print(df)
Output:
0 1
0 asdf 333
1 asdf 444
2 asdf 555
Default
read_csvseparator issep=r','.
Space as separator
Let’s now use the same function but space as a separator.
import pandas as pd
import io
temp=u"""
NSW VIC
6718023 5023203
6735528 5048207
6742690 5061266
6766133 5083593
6786160 5103965"""
df=pd.read_csv(io.StringIO(temp),
sep=r'\s{1,}', # one or more spaces
engine='python',
encoding = "iso-8859-1")
print(df)
Output:
NSW VIC
0 6718023 5023203
1 6735528 5048207
2 6742690 5061266
3 6766133 5083593
4 6786160 5103965
More than 1 spaces as separator
The next example will have NEW YORK as the index name, however, this should still be the single index name.
Example:
import pandas as pd
import io
s=u""" one two three four
INDIANA 0 1 2 3
COLORADO 4 5 6 7
NEW YORK 8 9 10 11
"""
df =pd.read_csv(io.StringIO(s),
sep=r'\s{2,}', # one or more spaces
engine='python',
encoding="iso-8859-1")
print(df)
The solution here is to use two spaces as a separator.
Output:
one two three four
INDIANA 0 1 2 3
COLORADO 4 5 6 7
NEW YORK 8 9 10 11
In here the dataframe index names are:
- INDIANA
- COLORADO
- NEW YORK
New line as a separator
Very similar example, but now, we use the new line as a separator.
Example:
from io import StringIO
import pandas as pd
text="""The lion (Panthera leo) is a species in the family Felidae.
It is a muscular, deep-chested cat with a short, rounded head.
A reduced neck and round ears, and a hairy tuft at the end of its tail.
The lion males are larger than females.
A typical weight range of 150 to 250 kg (330 to 550 lb) for males and 120 to 182 kg (265 to 400 lb) for females."""
df =pd.read_csv(StringIO(text),
sep=r'\n',
header=None,
engine='python',
encoding = "iso-8859-1",
names=["cname"])
print(df)
print("size:", df.size)
Output:

error_bad_lines=False
One special option to read bad lines, would be error_bad_lines=False.
Example:
import pandas as pd
import io
s=u""" one two three four
INDIANA 0 1 2 3
COLORADO 4 5 6 7
NEW YORK 8 9 10 11
"""
df =pd.read_csv(io.StringIO(s),
sep=r'\s{1,}', # one or more spaces
engine='python',
error_bad_lines=False,
encoding = "iso-8859-1")
print(df)
Here the NEW YORK row will fail, because it has an extra column. If we use the error_bad_lines=False option we will ignore this problem.
Output:
one two three four
INDIANA 0 1 2 3
COLORADO 4 5 6 7
Skipping line 4: Expected 5 fields in line 4, saw 6. Error could possibly be due to quotes being ignored when a multi-char delimiter is used.
When the load is slow
Suppose you have 100 million rows inside your DataFrame. You may run out of memory unless you set the dtype parameter:
dtype: Type name or dict of column -> type, optional
Data type for data or columns. E.g. {'a': np.float64, 'b': np.int32, 'c': ‘Int64’} Use str or object together with suitable na_values settings to preserve and not interpret dtype. If converters are specified, they will be applied INSTEAD of dtype conversion.
Because pandas will try to understand the column type by analyzing all the data inside any column.
In other words, by setting the column types within read_csv you will save some memory.
Parse as a date type
When reading using read_csv method in case pandas cannot recognize the data type columns we may set the following:
df = pd.read_csv(f'{path}file.csv',
sep=r',',
parse_dates=['dc1', 'dc2'])
In here the dc1 and dc2 are column names.
Read dataframe from the CSV file
Another approach would be to read the CSV on disk file.
Again we will use read_csv function which is almost identical as the read_table function. The only difference is:
read_csvuses comma separatorread_tableuses tab as separator
Usually the code to read the csv file will be short as this:
import pandas as pd
dataframe = pd.read_csv("test.csv")
Problems when loading csv files:
First if there is no file you may get the FileNotFoundError. Frequent problem is the UnicodeDecodeError, where you should first understand the encoding of your file. To get the encoding of a file on Linux you may run file -i command and use that. If you think the things will work with the utf-8 encoding, you may use this trick:
- Read the text from a file
- Decode the text with
text.decode('utf-8')
Usually utf-8 encoding, should cover the non-standard characters. If this doesn’t work try with the utf-16 encoding.
To fix the encoding problems you may use your editor and save the csv file to particular encoding.
In some cases you may get the parsing errors. If this is the case, use the engine=’python’ option.
Read dataframe from remote csv file
Again, the same read_csv function works. This time we will use the requests package.
Check here -> how to use requests package.
Example:
import pandas as pd
import io
import requests
url='https://programming-review.com/wp-content/uploads/cities.csv'
s=requests.get(url).content
c=pd.read_csv(io.StringIO(s.decode('utf-8')))
Output:
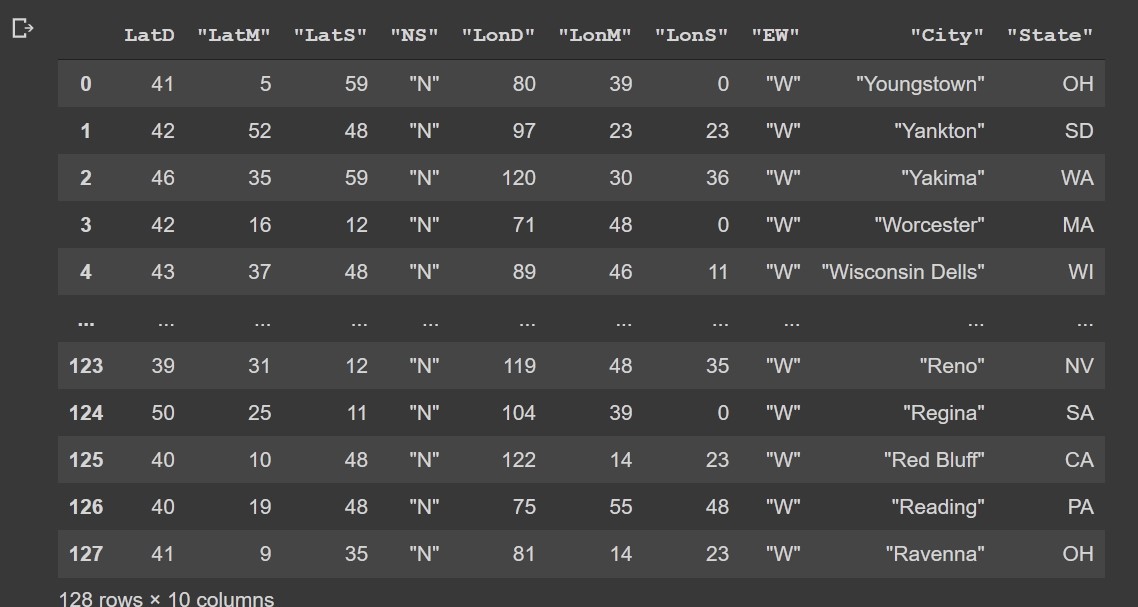
Read dataframe from HTML page
If there is a table (<table>) on a web page, pandas can read that. Note, this doesn’t work for divs.
Example:
import pandas as pd
df = pd.read_html('https://en.wikipedia.org/wiki/Comparison_of_web_browsers')[0]
df
This is equivalent to:
url = 'https://en.wikipedia.org/wiki/Comparison_of_web_browsers'
html_data = requests.get(url)
df = pd.read_html(html_data.text)[0]
df
Output:
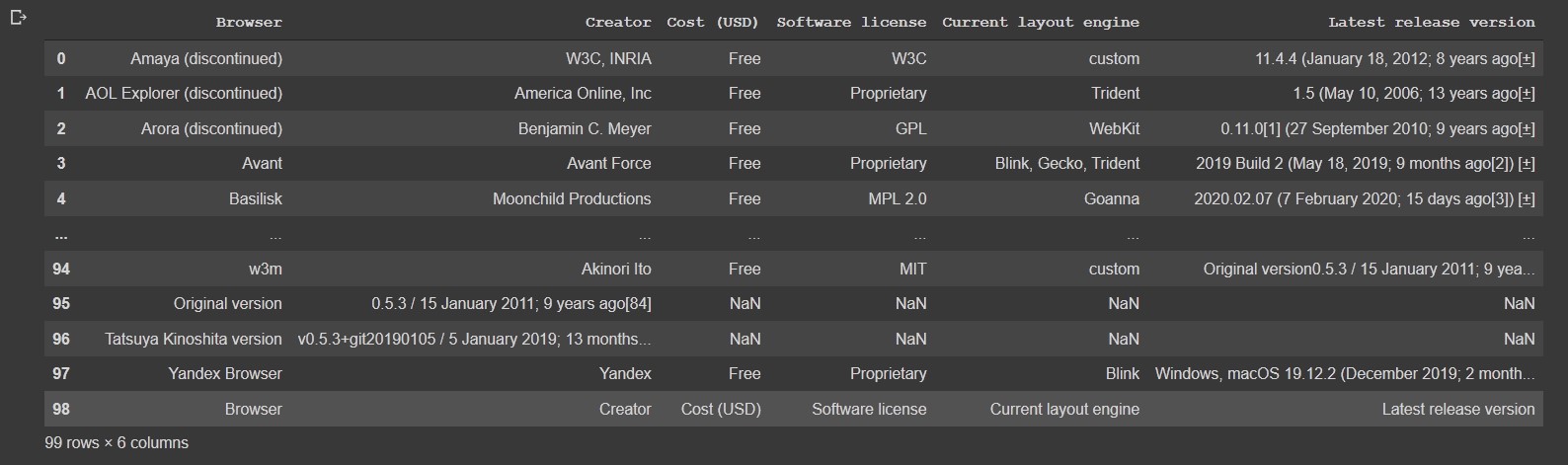
Fixing missing values
At times, you may have the missing values for some columns. It depends on a column, but often use this command:
df['col'].fillna((df['col'].median()), inplace=True)
In here column col should be a numeric type. This can be done even more robust:
Example:
from pandas.api.types import is_string_dtype, is_numeric_dtype, is_categorical_dtype
import os
import numpy as np
df = pd.DataFrame({'col1' : [1, np.nan, 3], 'col2' : ['elementary school', 'high school', 'middle school']})
def add_missing(df, cn):
if is_numeric_dtype(df[cn]) and df[cn].isnull().sum():
df[cn+'_na']=df[cn].isnull()
df[cn].fillna(df[cn].median(), inplace=True)
add_missing(df ,'col1')
df
Output:
col1 col2 col1_na
0 1.0 elementary school False
1 2.0 high school True
2 3.0 middle school False
Output:
col1 col2
0 1.0 elementary school
1 2.0 high school
2 3.0 middle school
While at first we had NaN special value.
col1 col2
0 1.0 elementary school
1 NaN high school
2 3.0 middle school
Convert string to categories
Often when we load data we like to convert string types to categories.
Example:
from pandas.api.types import is_string_dtype, is_numeric_dtype, is_categorical_dtype
df = pd.DataFrame({'col1' : [1, 2, 3], 'col2' : ['a', 'b', 'c']})
for k,v in df.items():
if is_string_dtype(v): df[k] = v.astype('category').cat.as_ordered()# as_unordered
df.col2.cat.categories
Output:
#Index(['a', 'b', 'c'], dtype='object')
With this process we converted the col2 column into type category from type object. df.info() yields:
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 3 entries, 0 to 2
Data columns (total 2 columns):
col1 3 non-null int64
col2 3 non-null category
dtypes: category(1), int64(1)
memory usage: 259.0 bytes
Before it was:
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 3 entries, 0 to 2
Data columns (total 2 columns):
col1 3 non-null int64
col2 3 non-null object
dtypes: int64(1), object(1)
memory usage: 176.0+ bytes
If you print the df you cannot change the difference, because the output will be the same in both the cases.
The change looks small, but beneath in pandas the difference is that we may now use machine learning algorithms on the non-string data.
To check some other forms of converting string data to numerical formats you may check the Encode Categorical Features article.
It is also possible to set your order of categories like this:
Example:
from pandas.api.types import is_string_dtype, is_numeric_dtype, is_categorical_dtype
df = pd.DataFrame({'col1' : [1, 2, 3], 'col2' : ['elementary school', 'high school', 'middle school']})
for k,v in df.items():
if is_string_dtype(v): df[k] = v.astype('category')
df.col2.cat.set_categories(['elementary school', 'middle school', 'high school'], ordered=True, inplace=True)
df.col2.cat.categories
Output:
Index(['elementary school', 'middle school', 'high school'], dtype='object')
Else the output would be alphabetical which is not what we may plan.
What machine learning algorithms need?
What the machine learning algorithms do not expect are raw strings. The upper method of conversion to category astype(‘category’) will at the end provide code:
Example:
from pandas.api.types import is_string_dtype, is_numeric_dtype, is_categorical_dtype
df = pd.DataFrame({'col1' : [1, 2, 3], 'col2' : ['elementary school', 'high school', 'middle school']})
for k,v in df.items():
if is_string_dtype(v): df[k] = v.astype('category')
df.col2.cat.set_categories(['elementary school', 'middle school', 'high school'], ordered=True, inplace=True)
df.col2.cat.codes
Output:
0 0
1 2
2 1
dtype: int8
where in the column to the right 0 stands for elementary school, 1 stands for the middle school and 2 for high school.
Fast save and load DataFrame in pandas
When dealing with big DataFrames (100M rows) you may find to_feather and read_feather as an excellent choice:
Example:
os.makedirs('tmp', exist_ok=True)
df.to_feather('tmp/row') # !pip install pyarrow
To load the data you use:
Example:
df=pd.read_feather('tmp/row')
df
This actually reads pandas memory and saves it to a disc and reads from disk to pandas memory.
…
tags: pandas - string - load & category: python
